Background
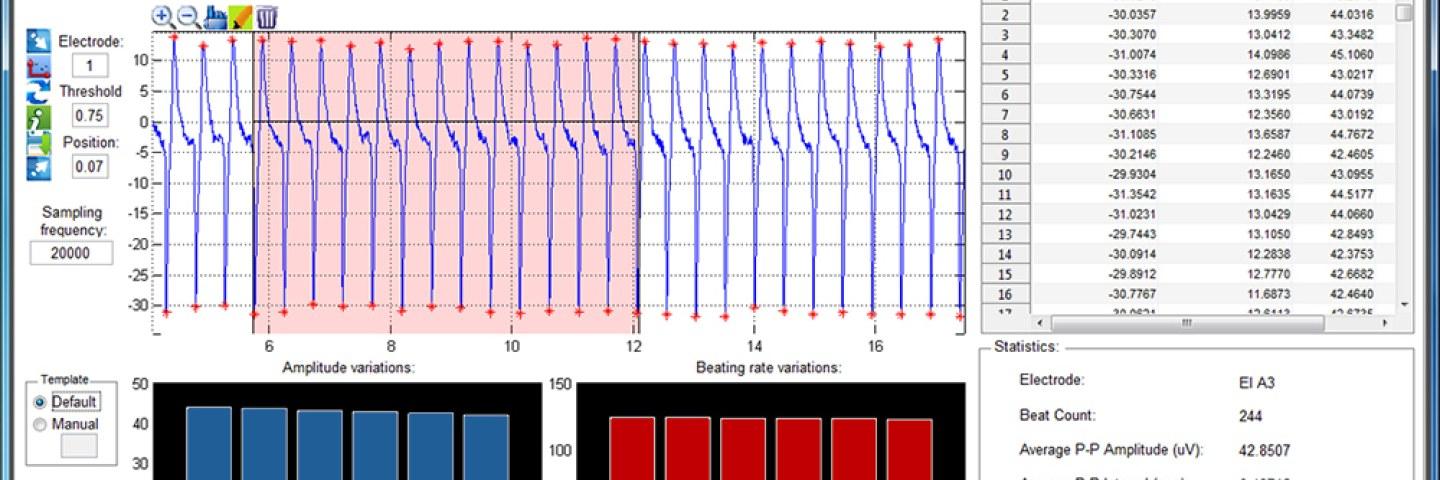
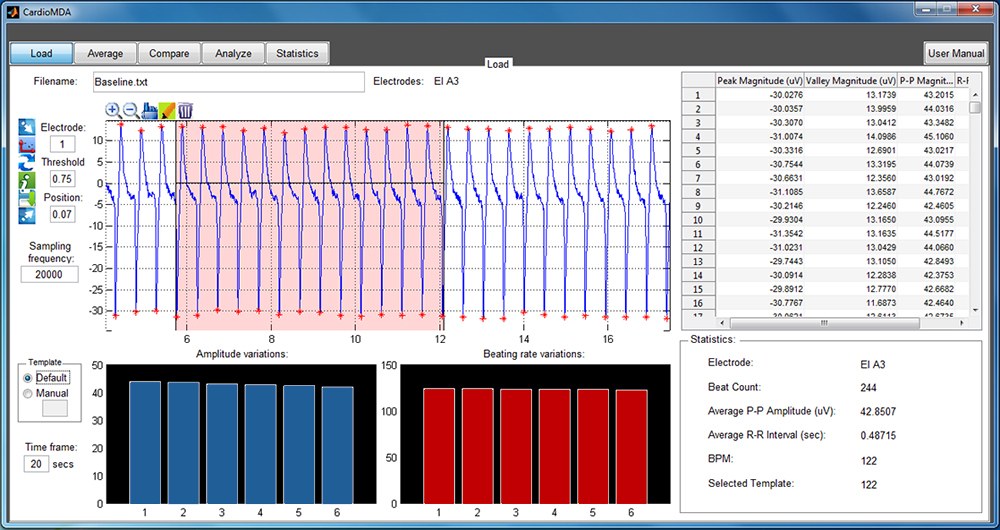
Cardiomyocyte MEA Data Analysis (CardioMDA) software is a simple and semi-automated tool to analyze and evaluate drug response in cardiomyocyte cell population from field potential recordings. This offline analysis tool is designed to improve accuracy and reliability of drug screening. The software requires no prior programming skills and it’s easy to use interface allows researchers and clinicians in different laboratory settings to utilize the software. The batch processing utility and automated customization for parallel processing is an advantage, especially when huge datasets need to be analyzed.
The program is distributed as a self-extracting package with the MATLAB Compiler Runtime Version 8.1 that needs to be installed for the program to work. The user manual, sample data and other files needed for running the software are provided in the package.
Please note that registration is required before access to the download package, and this analysis software is for non-commercial purposes and research use only.
Features of the CardioMDA software
CardioMDA was designed to work for data recorded on Multichannel system (MCS) amplifiers. The current version of the software is only compatible to custom ASCII file formats with suitable headers to identify electrodes, exported using MC_DataTool software. Other file formats and customizations may be considered in the future versions.
Some other notable features of the CardioMDA software are:
- An easy to use and self-intuitive graphical user interface.
(Check out screenshots on the right.) - Customizable parameters to account for different signal morphologies and quality.
- Correlation analyses and ensemble averaging to identify and eliminate sporadic or arrhythmic beats.
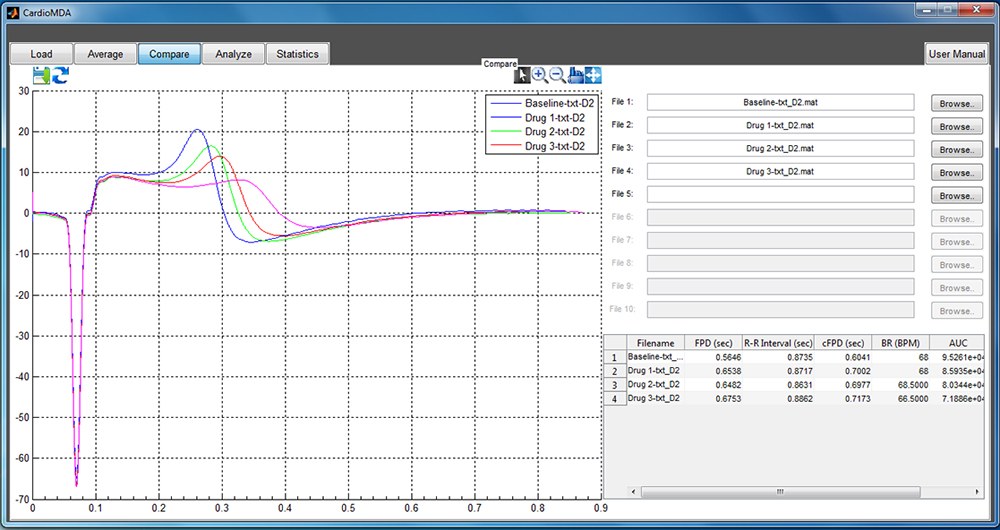
- Overlay plots and comparison tables for up to 10 different signal sources.
- Batch processing utility with automated customization for parallel computing*
to minimize processing times and user interaction. - Available in both 32-BIT and 64-BIT versions.
* Parallel computing is configured only for systems running on multi-core processors.
Feedback and suggestions are welcome
The current version (Version 1.0) is at the moment under beta-testing phase. However, it may be distributed upon request to enable us to collect user feedback and suggestions on ways how the software could be improved and provide flexibility in the analysis. With your feedback and suggestions, new features may be included in the future versions
Support
If you have any trouble or questions regarding CardioMDA that are not addressed in the User Manual please send us email at cardiomda(at)lists.tuni.fi. The software is a beta version and under development thus any feedback and questions are valuable.
The software takes about 1 - 1.5 minutes to load in the first instance after start-up.
Reference article:
Cardiomyocyte MEA Data Analysis (CardioMDA) – A Novel Field Potential Data Analysis Software for Pluripotent Stem Cell Derived Cardiomyocytes
Abstract
Cardiac safety pharmacology requires in-vitro testing of all drug candidates before clinical trials in order to ensure they are screened for cardio-toxic effects which may result in severe arrhythmias. Micro-electrode arrays (MEA) serve as a complement to current in-vitro methods for drug safety testing. However, MEA recordings produce huge volumes of data and manual analysis forms a bottleneck for high-throughput screening. To overcome this issue, we have developed an offline, semi-automatic data analysis software, ‘Cardiomyocyte MEA Data Analysis (CardioMDA)’, equipped with correlation analysis and ensemble averaging techniques to improve the accuracy, reliability and throughput rate of analysing human pluripotent stem cell derived cardiomyocyte (CM) field potentials. With the program, true field potential and arrhythmogenic complexes can be distinguished from one another.
The averaged field potential complexes, analysed using our software to determine the field potential duration, were compared with the analogous values obtained from manual analysis. The reliability of the correlation analysis algorithm, evaluated using various arrhythmogenic and morphology changing signals, revealed a mean sensitivity and specificity of 99.27% and 94.49% respectively, in determining true field potential complexes. The field potential duration of the averaged waveforms corresponded well to the manually analysed data, thus demonstrating the reliability of the software.
The software has also the capability to create overlay plots for signals recorded under different drug concentrations in order to visualize and compare the magnitude of response on different ion channels as a result of drug treatment. Our novel field potential analysis platform will facilitate the analysis of CM MEA signals in semi-automated way and provide a reliable means of efficient and swift analysis for cardiomyocyte drug or disease model studies.
Read the whole article: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0073637