New GeoMx device allows detailed spatial characterisation of tissue samples at Tampere University
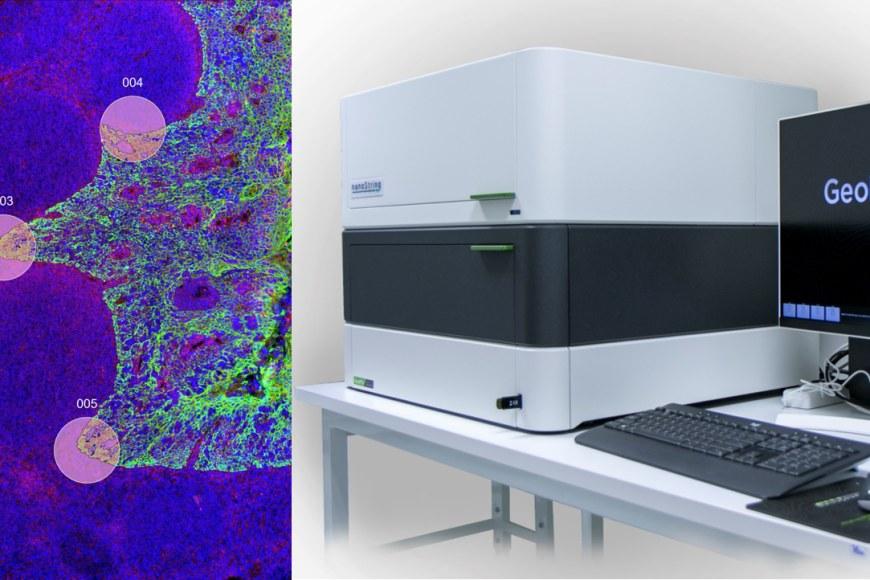
Tampere University has acquired a new device from NanoString, the GeoMx Digital Spatial Profiler, which will make spatial omics available to researchers in Tampere and Finland. The investment was initially promoted by adjunct professor Kirsi Granberg, PI of the Cancer Regulation and Immunology research group.
Spatial transcriptomics, one of the major applications of the profiler, was chosen as the method of the year 2020 by the Nature journal. It generates possibilities to analyse the state and behaviour of, for example, cancer cells, immune cells, or other cells in the pathological tissue so that their location and proximity to each other is known, which helps to understand the development of diseases, and treatment responses.
“This is also clinically relevant because some known drug resistance mechanisms are cell type-specific but also dependent on the cell-cell interactions with the surrounding cells,” says Postdoctoral Research Fellow Alejandra Rodriguez Martinez who coordinates the use of the new device together with doctoral student Aliisa Tiihonen.
“Only spatially resolved data contains the information required to identify these types of mechanisms and select effective therapies for the patients,” Rodriguez Martinez adds.
GeoMx allows selecting specific tissue compartments or cell types for analysis through morphology marker visualisation. After selection, an ultraviolet light is very precisely directed to each of the selected areas, resulting in the release of UV-cleavable oligomers, which carry the molecular information needed to subsequently quantify the presence of different transcripts or proteins in the sample.
“Most importantly, the method allows linking transcript or protein levels to the different areas or cell types in the sample,” Tiihonen says.
Both fresh frozen and Formalin Fixed Paraffin Embedded (FFPE) samples can be used in the analysis.
The GeoMx device is in the Faculty of Medicine and Health Technology. It is part of the Histology Laboratory in the Arvo building on the Kauppi campus. Internal users may use the device after training and other users are offered the possibility to have their samples processed as a service, done by laboratory assistant Päivi Martikainen.





